MaxFidelity Taq DNA Polymerase
Catalog Number: TMFV100RX
Unit Size: 100 rxns (1ML)
Description
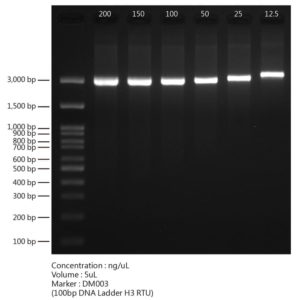
MaxFidelity Taq is a new recombinant enzyme with genetic modification for its amino acid sequence, which results 70 times better fidelity than Taq DNA polymerase and an extremely fast elongation rate (as fast as 15 seconds per kilo base, kb). MaxFidelity Taq has higher stability at high temperature, and this property makes MaxFidelity Taq perform very well for GC-rich PCR. Being highly thermo-stable, MaxFidelity Taq DNA Polymerase can remain viable even after being subjected to boiling for 2 minutes. This special enzyme has been modified genetically and needs less concentration of magnesium than other polymerases. The suggestion for magnesium ion in the reaction is 0.8 to 1.2 mM. 10X MaxFidelity Taq PCR buffer contains no magnesium. A tube of 25 mM MgSO4 is provided. Further optimization can be achieved for different targets of DNAs. Reagents are provided for 100 PCR reactions of 50 μl each.
Features:
- 5’→3’ DNA polymerase activity
- High reaction rate: 10 seconds/kb.
- Generates blunt end amplicon.
- 3’→5’ exonuclease (proofreading) activity.
- High fidelity: 70 times higher than Taq polymerase.
Kit Contents:
| Contents | MB601-0100 | MB601-0010 |
| MaxFidelity Taq DNA Polymerase (1 U/μl) | 100 μl x 1 vial | 10 μl x 1 vial |
| 10X MaxFidelity Taq PCR buffer | 1 ml x 1 vial | 100 μl x 1 vial |
| 25 mM Magnesium Sulfate | 1 ml x 1 vial | 100 μl x 1 vial |
| dNTP Mix (2mM each) | 1 ml x 1 vial | 100 μl x 1 vial |
| DMSO | 1 ml x 1 vial | 100 μl x 1 vial |
Quality Control
The quality of the MaxFidelity Taq DNA Polymerase is tested on a lot-to-lot basis to ensure consistent product quality.
Storage Buffer of MaxFidelity Taq DNA Polymerase
50 mM Tris-HCl (pH 8.1), 60 mM KCl, 0.1 mM EDTA, 1 mM DTT, stabilizers, and 50% (v/v) glycerol.
Unit Definition
One unit of MaxFidelity Taq DNA Polymerase incorporates 10 nmol of deoxyribonucleotide into acid-insoluble material in 30 minutes at 74°C.
Template
MaxFidelity Taq DNA Polymerase is suitable for amplifying targets up to 10 kb from the following templates:
| Template | Amount |
| Genomic DNA | 10-200 ng |
| Plasmid DNA | 1-5 ng |
| cDNA | ~100 ng starting total RNA |
Amplification of longer targets (up to 10 kb) may be possible, but may require more template and longer elongation times.
Primers
Use 0.3 μM per primer as a general starting point. For larger amounts of template (e.g., 200 ng genomic DNA), increasing the concentration up to 0.5 μM per primer may improve yield.
Annealing Temperature
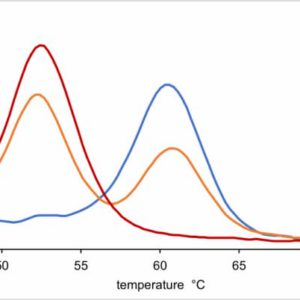
The annealing temperature is slightly higher than with typical PCR. The optimal annealing temperature should be ~2°C lower than the Tm (melting temperature) of the primers used. A range of 58-68°C is recommended. MgSO4: MgSO4 is not included in the 10X MaxFidelity Taq PCR buffer, and we suggested working concentration is 1 mM, which is sufficient for most templates. For further optimization, modify the amount of MgSO4 for final 0.8-1.2 mM of the concentration.
Extension Time
As little as 15 seconds per kb may be used; 30 seconds per kb is suitable for most targets.
We recommended use up to 60 seconds per kb for maximum yield.
Required Materials
- PCR tube
- PCR instrument
- RNase-free H2O
- DNA electrophoresis equipment
MaxFidelity Taq DNA Polymerase Protocol
- For each 50 μl reaction, assemble the following components in a 0.2 ml PCR tube on ice before the experiment:
| Component | Volume (μl) | Final Concentration |
| 10X MaxFidelity Taq PCR Buffer | 5 μl | 1X |
| 2 mM dNTP Mix | 5 μl | 0.2 mM each |
| 2 mM MgSO4 | 2 μl | 1 mM |
| Primer mix (10 μM each) | 1.5 μl | 0.3 μM each |
| Template DNA | Variable | see Template Part |
| MaxFidelity Taq DNA Polymerase (1 U/μl) | 1 μl | 1 U |
| DMSO (for GC-rich target) | Variable | 5~10% |
| Autoclaved, distilled water to | 50 μl | |
- Mix gently. If necessary, centrifuge briefly. Cap the tube and place it in the thermal cycler.
- To process in the thermal cycler for 35 cycles as follows:
| Process | Temperuture (°C) | Time | Cycles |
| Initial Denaturation | 96-98 | 2 minutes | 1 |
| Denaturation | 94 | 15 seconds | 35 |
| Annealing | 60-68 (Tm of primers minus 2°C) | 10-30 seconds | |
| Extension | 68 | 30-60 seconds per kb | |
| of PCR product | |||
| Final extension | 68 | 5 minutes | 1 |
Note: Optimal conditions for amplification will vary depending on the primers and thermal cycler used.
It may be necessary to optimize the system.
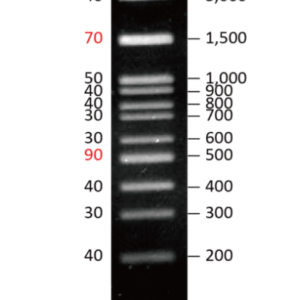
- After the PCR reaction, analyze products using standard agarose gel electrophoresis.
Troubleshooting
Refer to the table below to troubleshoot problems that you may encounter when you did PCR amplification with the kit.
| Problem | Cause | Solution |
| Low yield of | Incomplete concentration | Use the appropriate method for the DNA preparation |
| PCR products | of start materials | based on the amount of the starting materials. |
| DNA degrade | DNA is not fresh | Avoid repeated freeze / thaw cycles of the sample. |
| Keep DNA preparations on ice or frozen in order to | ||
| avoid the degradation. | ||
| DNase contaminant | Use the fresh TAE or TBE electrophoresis buffer. | |
| Maintain a sterile work environment to avoid | ||
| contamination from DNase. |
Caution
- During operation, always wear a lab coat, disposable gloves, and protective equipment.